Training pixel classifiers
While we were running the 2023 From Samples to Knowledge QuPath workshop, a common question we received was why we discourage the use of large area annotations when training a pixel classifier.
Not only does using large area annotations saturate the model with very similar types of pixels (many more “central” pixels than “edge” pixels), the large amounts of data users have already fed into the model make it very difficult to fine tune. Imagine trying to nudge an elephant into the right position for a photo rather than a cat or a dog - momentum and mass matters! Classifiers trained with massive numbers of pixels will be difficult to adjust, and the situation is worse, in fact, than a simple matter of overcoming a mass of training data.
While the above comments apply generally to any kind of classifier you supply with training data, whether it be object or pixel, in QuPath or otherwise, QuPath has yet another setting that tends to fly under the radar that impacts this discussion. Two, in fact. Both are tucked away within the Advanced Options menu shown below.
While there are a variety of options here, the two I want to highlight are Maximum samples and Reweight samples.
Maximum samples - The mouseover help fairly straightforwardly describes it as the maximum number of training samples allowed, but does not go into a lot of details otherwise. What “samples” means in this case are individual pixels. When you have more than the maximum number of samples, QuPath will use the Random seed to “randomly” (except that it will be the same pixels every time, for a given set of pixels, as long as the seed is kept constant) select a subset of those pixels.
Reweight samples - This checkbox allows QuPath to weight the subsets of the total 100,000 pixels being used for training by the class balance. If you have 80% of your data as one class, and 20% as another, it will give more weight - more impact within the classifier - to the less frequent class based on the relative lack of data for that class or classes. It is a way of attempting to ensure fairness within a classifier where the the training data provided is unfair.
 Warning
Warning
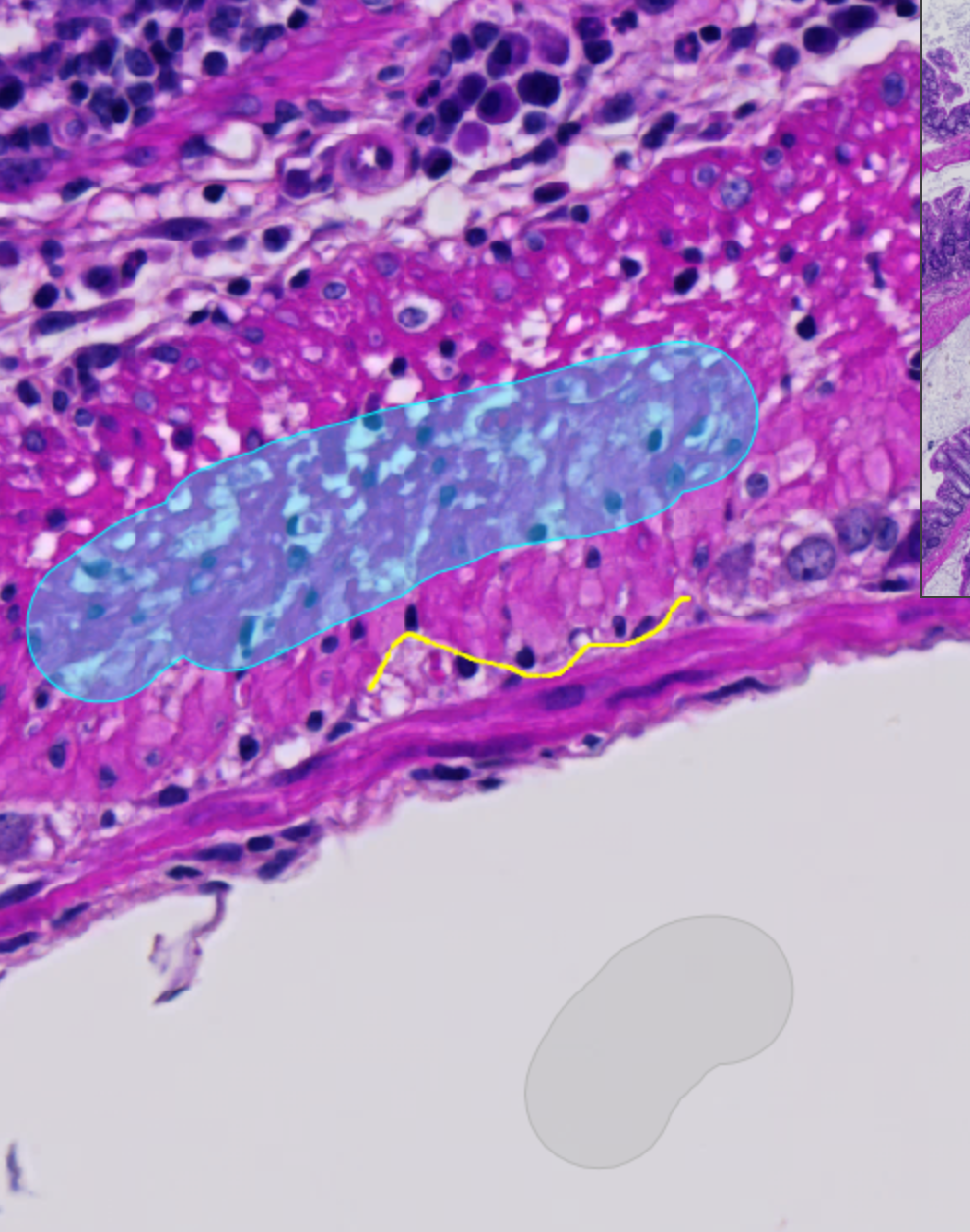
Where am I going with this in relation to generating training data with the wand or brush tools? Let’s take a small example where I have created a simple pixel classifier with some training data using the brush tool. See the images below.
The setting is that the “Other” and “Ignore*” classes were the first two classes I trained, but I found that the Other class was not doing a very good job around the borders. To try and fix this, I try to generate some training data around the edges with a line tool. For the purposes of this example, the line is highlighted in yellow in the picture on the left, and I chose the classification to be “Necrosis” rather than Other so that it would show up separately in the pie chart. Even so, you can barely see the Necrosis part of the pie chart, pointing directly down in the image.
When you use large areas as the basis of your training data, it becomes very difficult to train the classifier to pay attention to small things like the edges, which are actually one of the most important things to you! There will almost always be many “obvious” pixels around the center of a type of tissue that are very easy to annotate quickly, but you do not want those to obscure/hide/drown the data that is most important to your classifier, that which defines the border regions. In the end, we care most about those edges, because they truly define and contain the areas we are interested in.
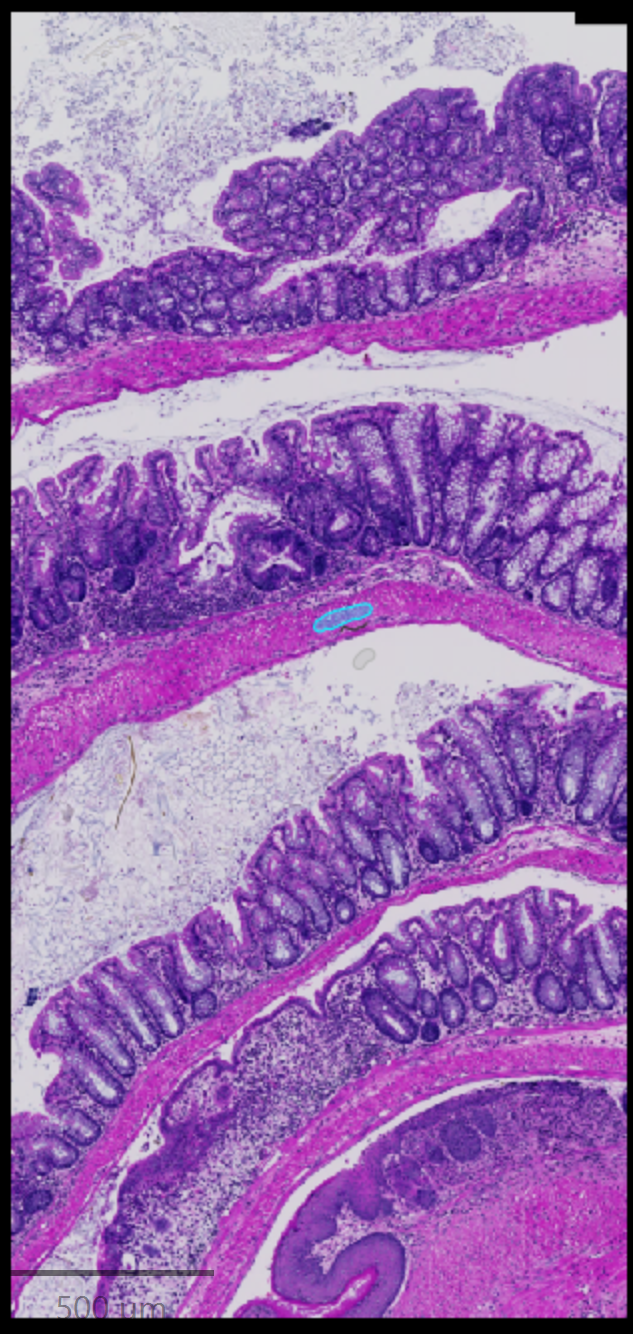
In QuPath at least, and probably most other software, it gets worse, and this brings us back to the Advanced options dialog we were looking at before. In order to prevent memory problems or slowing the classifiers down to a crawl, most classification programs or scripts will take a subset of the data as “representative”, and use that to perform the analysis. In the specific case we were looking at above, the annotated area seemed rather small, and here it is in context.
Zooming out slightly, and looking at the actual area used for training “Other”, it does not look like very much. In fact the full image is shown on the far right, and the annotated area is barely visible near the middle of the image. However, this ~5000 square micron area contains approximately 370,000 pixels (4883/(0.111*0.111)).
But wait, our pixel classifier only handles 100,000 pixels! Of ALL classes combined. That means we are not actually benefiting from that large training area, and if we attempt to add small refinements to it, there is a chance we will not even use the training data we added. Now, since the added training data was shown as Necrosis, class balancing could help make up for that somewhat… but that training area was not actually intended to be Necrosis, that was only to demonstrate within the pie chart - that tiny sliver of the pie chart that was so important for defining the border areas would actually have been washed away in the giant sea of pixels we started with if correctly annotated as Other.
Let us take a look at how this could have looked using line annotations as inputs.
Here I made the line width as shown by QuPath slightly thicker so that the lines themselves would be more visible, but this does not change the number of pixels that are used for training data - the lines are still lines between points and do not have width. The fake “Necrosis” line is the same size as previously, but now the pie chart shows that it makes up a significant percentage of the total data, and will therefore have more of an impact on the final classifier. In summary, it will be far easier to adjust your classifiers if you use smaller amounts of pixel training data, especially when refining the pixel classifier is important.
 Info
Info
Empty