Command line and Python
Work in progress, will keep updating as I have time and a project to bring me back to using Python
 Note
Note
Using the command line on its own
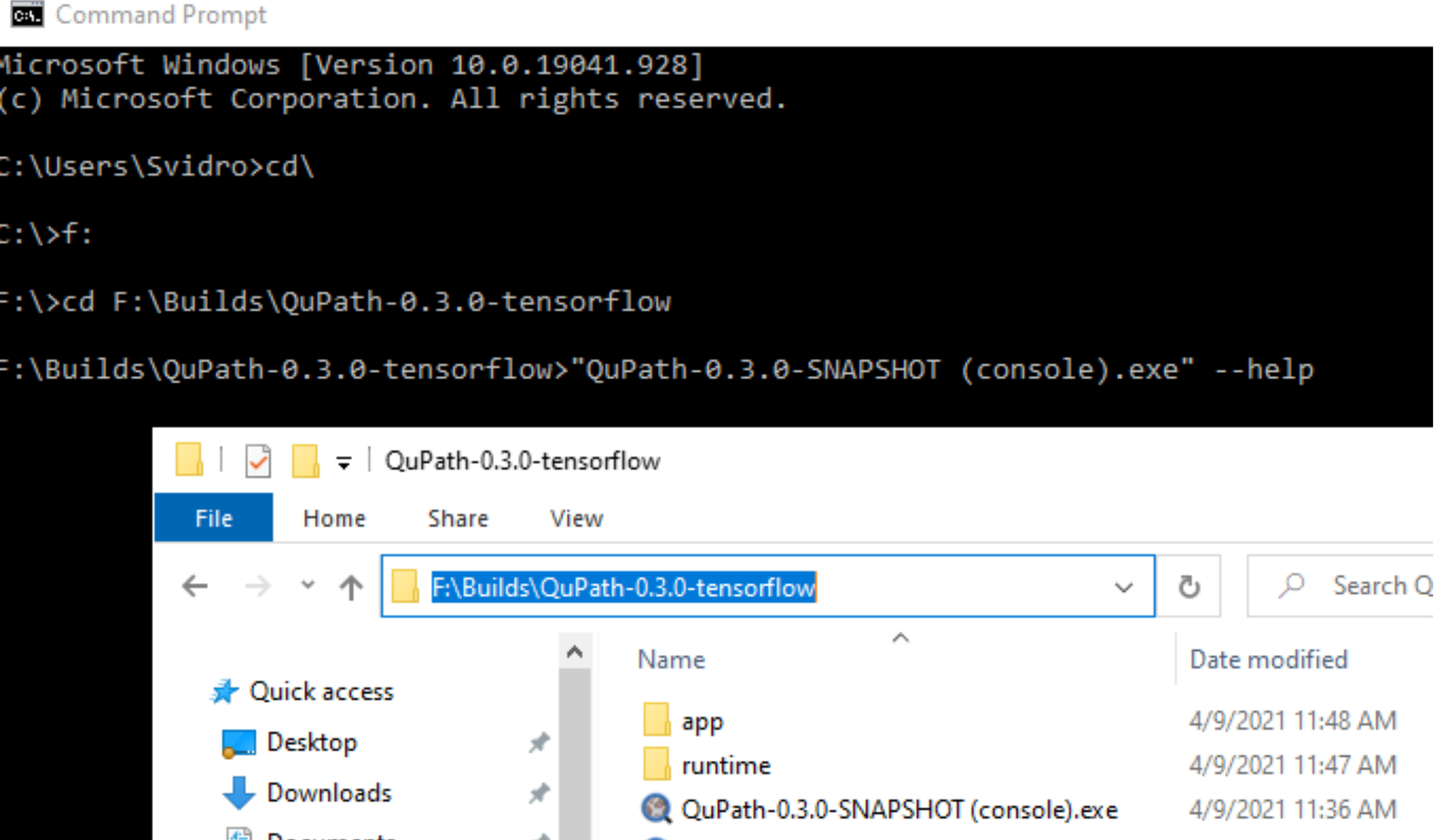
Navigate in a command prompt to the QuPath installation directory, and follow the same steps as listed above.
Here I navigated to the directory where the QuPath executable is, and added in the “- -help” flag
In my case I used "F:" to navigate to the F drive where my QuPath installation existed, then copied the directory path from the bar at the top into a "cd " command, and hit enter.
Then I copied the text of the file name into a pair of quotation marks, and added --help onto the end. Pressing Enter gets me the help information.
To run a script like the following, I can pass an image and the script in the following format:
"QuPath-0.3.0-SNAPSHOT (console).exe" script --image=f:\InputImages\1.tif f:\dummy.groovy
The order is important!
Here is a quick dummy script that has two different ways of printing out the file name, pick whichever one works for your image type.
//dummy.groovy //print getCurrentServerPath() println("Script starts") //I found that the above fails for the type of image I initially tested the script on, and I needed to use path= getCurrentServer().getBuilder().getURIs()[0].toString() print path +"\n" //Maybe do some other things here like convert the image type def pathInput = path.substring(6,) println(pathInput) println("The script ran and printed the file location")
After running dummy.groovy, the file name and a line of text is printed. Not useful, but maybe enough to get someone started.
 Info
Info
Accessing the command line from WITHIN a Groovy script
In my case usually to run Python code, you can use something like:
// Get the imageData & server def imageData = getCurrentImageData() def server = imageData.getServer() String path = server.getPath() def cmdArray = ["python", "c:\\Python\\Test1\\Email alert.py", path] cmdArray.execute()
where Email alert.py is a Python script file, and the first argument is the command I would use on the command line to run that script. “path” is included as an argument that the Python script will have access to (sys.argv[1]) below.
In general, the script would run as if the command were typed into the command line in this way:
 Warning
Warning
#This script was largely taken from Stackoverflow and is intended to be called from within #QuPath in order to alert the "recipient" email address that a particular slide had finished processing import smtplib import sys from email.mime.multipart import MIMEMultipart from email.mime.text import MIMEText #free to use spare email account gmailUser = 'qupathemailer@gmail.com' gmailPassword = 'no longer a password' recipient = 'targetEmail@domain.com' message = 'QuPath script has completed on '+sys.argv[1] msg = MIMEMultipart() msg['From'] = gmailUser msg['To'] = recipient msg['Subject'] = "QuPath Alert" msg.attach(MIMEText(message)) mailServer = smtplib.SMTP('smtp.gmail.com', 587) mailServer.ehlo() mailServer.starttls() mailServer.ehlo() mailServer.login(gmailUser, gmailPassword) mailServer.sendmail(gmailUser, recipient, msg.as_string()) mailServer.close()
Converting images to OME-TIFF via command line
A more complete example of interacting with Python shown below involves using Python to call QuPath from the command line repeatedly to convert the provided image into another type, and pyramidalize it.
Below is a Python script that uses a Groovy script found at a particular location, indicated by the variable script. The workflow would be, after having QuPath and Python set up, to run this script targeting the InputImages folder and have it process all images within. Alternatively, the script could be run persistently and “watch” the imageDirectory for new files.
See the first Note on this page for a script that can convert multiple files to OME.TIF from within QuPath.
import os import subprocess import time script = "F:\\Scripts\\Export OMETIF from CLI v2.groovy" imageDirectory = "F:\\InputImages\\" outputDirectory = 'F:\\ProcessedImages\\' qupath = "F:\\Builds\\QuPath-0.2.3\\QuPath-0.2.3 (console).exe" fileList = os.listdir(imageDirectory) for file in fileList: if (file.endswith('.tif') and not file.endswith('.ome.tif')): imageFile = imageDirectory + file outputImage = outputDirectory + file subprocess.run([qupath, "script", script, "-i", imageFile], shell=True) #subprocess.run([qupath, "convert-ome", imageFile, outputImage, "-y=4", "-p"], shell=True) os.rename(imageFile, outputImage)
Roughly speaking, the above script will take images in one folder (a fileList of everything in imageDirectory), convert anything with the .tif extension to OME.TIFF, and save them in a new folder.
The images this was tested on were single channel TIFF images, so other steps may need to be taken for particular formats and data types. The script needs to know where the QuPath executable file is so that it can make use of that, and then either uses a script designed to perform the conversion, or, currently commented out, uses QuPath’s command line to write a pyramidal (-p), 4 layer (-y=4) image.
More information should be available using the “--help” flag on the command line as described at the top of the page.
// source https://forum.image.sc/t/issue-with-bfconvert-and-big-jpg-images/43276/2 // and https://forum.image.sc/t/using-qupath-to-convert-very-large-tif-to-ome-tif-and-add-metadata/48636/4 // With grand struggles from Mike Nelson // QuPath 0.2.3 server = getCurrentServer() // I found that server.getFile() fails for this type of image, and I needed to use // For other file types, this might not all be necessary. path= server.getBuilder().getURIs()[0].toString() imageData = getCurrentImageData() baseFilePath = path.substring(6, path.lastIndexOf(".")+1) def pathInput = baseFilePath + "tif" def pathOutput = baseFilePath + "ome.tif" println 'Reading image...' //def img = ij.IJ.openImage(pathInput).getBufferedImage() //def server = new WrappedBufferedImageServer("Anything", img) def metadataNew = new ImageServerMetadata.Builder(server.getMetadata()) .pixelSizeMicrons(1.16, 1.16) .zSpacingMicrons(1) .build(); imageData.updateServerMetadata(metadataNew); println 'Writing OME-TIFF' new OMEPyramidWriter.Builder(server) .parallelize() .tileSize(512) .scaledDownsampling(1, 4) .build() .writePyramid(pathOutput) println 'Done!' import qupath.lib.images.writers.ome.OMEPyramidWriter import qupath.lib.images.servers.* import javax.imageio.* import qupath.lib.images.servers.ImageServerMetadata;
Another, more interesting look at the interaction between Python and QuPath was done by BIOP, and is hosted here.
It involves importing ImageJ ROIs instead of masks, but the results should be similar.
Other links on the topic of the command line, Python, and QuPath and complex scripting
Calling a python .py file from within a QuPath script.
Directly importing the results of something run externally, say in Python. Specifically batch importing results from CellPose. A much better option is included next.
An extension for running CellPose on a whole slide image, with options to handle overlap resolution in tiles. This link is not particularly useful in terms of learning code, but it is a much more useful bit of code for making use of CellPose.
Calling scripts from other scripts, along with a warning about import statements!
Accessing different images within a multi-image file, for example macro images.