Tissue detection (simple thresholder)
Pixel classification (tissue regions)
Cell creation
The last measurement we need requires the creation of cells in various parts of the tissue. Cell detection in QuPath is primarily based on detection of the nucleus. That is not to say that cell detection cannot be used to detect other things - just that its purpose is based around finding an object of small round-ish object of a single stain or channel. In brightfield images, Optical Density can also be used when the nucleus could be multiple colors, and those colors obscure each other, as in the case of KI-67 staining with DAB.
 Note
Note
Preparing to test cell detection
First I run the script as far as we have gotten it, so that I can look at the state of the project as I begin cell detection. I will NOT run this in the training images as it will delete all of the training data!!! It would be safest at this time to back up your training data by saving a duplicate copy of the project or the .QPDATA files for those images. The image of the Hierarchy tab to the right shows what I have so far - one parent annotation (the tissue detection), and three child annotations indicating the different tissue types. The goal is to generate cells in the EosinDense and NormalTissue annotations.
If I were to select all annotations and run Cell detection, I would end up with a lot of cells within the original Tissue annotation, with all other annotations deleted. That is because once Cell detection starts within a given annotation, it removes all objects within that annotation. We need to specifically select the annotations we want to run Cell detection on, and version 0.2.0+ gives us an easy way to do that.
Selecting annotations by class: Go to the Annotations tab, and select the NormalTissue and EosinDense classes (SHIFT+click or CTRL+click to select multiple classes, like in a file folder). Right click for the context menu, and select the bottom option, Select objects by classification. The two annotations of those classes will now be selected, and in the Workflow tab, you should have a new entry that will let you perform this action by script.
selectObjectsByClassification("EosinDense", "NormalTissue");
Great! Now we are all set to run cell detection. I recommend manually creating a few small areas to test out different settings, and make these areas fairly varied in terms of cell density and stain type. Remember, we just saw in the Pixel classifying section what the increased cell density could do to class detection! At some point later I may go into a deeper dive on various cell detection methods, but for the moment I will simply leave a set of links with further information, and that it can be a nice trick to make several small square annotations quickly, select them all and merge them using the Objects menu, and then quickly iterate your cell detection on that distributed single annotation.
The main advantage of testing in small regions is really… your patience. If you have to wait 5-10 minutes per iteration, most people will get annoyed and tired of waiting, and will test fewer times. Testing in small regions is not an excuse to not check your results after running it on the full region, but it should allow you to quickly adjust variables and do a fairly good job of creating a useful cell detection algorithm in a short time. Provided the areas represent a good cross section of the different types of tissue areas.
Testing cell detection
 Cell Detection Resources
Cell Detection Resources
Several rectangular annotations merged into a single annotation in Tile 4 using Objects->Annotations->Merge selected
After a few quick tests, the only settings I changed were reducing the Maximum area to 200 (just in case of artifacts, none of the nuclei I found were even close), and increased the Hematoxylin OD Threshold to 0.25 to exclude some of the nuclei that were not fully in-plane and to reduce issues with adjacent HematoxylinDense tissue areas.
Copying from the workflow:
runPlugin('qupath.imagej.detect.cells.WatershedCellDetection', '{"detectionImageBrightfield": "Hematoxylin OD", "requestedPixelSizeMicrons": 0.5, "backgroundRadiusMicrons": 8.0, "medianRadiusMicrons": 0.0, "sigmaMicrons": 1.5, "minAreaMicrons": 10.0, "maxAreaMicrons": 400.0, "threshold": 0.25, "maxBackground": 2.0, "watershedPostProcess": true, "cellExpansionMicrons": 5.0, "includeNuclei": true, "smoothBoundaries": true, "makeMeasurements": true}');
Classification - Selecting training data
Since I wanted to include some classification, I went ahead and saved the data with the test annotations, and then duplicated this image (Tile 4, with data) to create a cell classifier training image. This training image is only really necessary since I am going to use a machine learning classifier to try to detect elongated cells, but normally if elongated cells were the only thing I were interested in, I would choose a single measurement classifier based on the Eccentricity.
 Warning - Selecting training data
Warning - Selecting training data
In the cell training image, I selected the Points tool (three circles in a triangle) which opened up the Counting dialog box. In the Counting dialog box I clicked Add twice to create two groups, then assigned each group a class (Positive and Negative in this case, although you can create classes for these just like for the pixel classifier) by right clicking and Set class. At this point, all I need to do is select one of the two Points objects and then click somewhere within a cell. That will drop a point of the selected class within that cell. Having a Point object within a cell during training lets QuPath know that you want that cell to be treated as a particular class. It does not guarantee that your classifier will end up giving that particular cell that class - only that it is to be treated as a training example of what you want that class to look like. Also, “look like” is probably a bad choice of words here, as the only thing the classifier will use are the cell measurements. The QuPath classifiers, at this time, never “see” the cell. Any measurements you might have added afterwards will be included with the default measurements generated when the cells are created. Any measurements added after the classifier is created will not be included or used.
Another, faster but less precise, way of generating training data is to use the Brush or other area annotation tool to select an area and then apply a class to that. All cells that have a centroid within the annotation (not simply intersecting with the annotatino) will be considered that annotation's class during training. Pete has an example in his YouTube playlist.
Current state of the project
Adding points to a training area.
 Warning
Warning
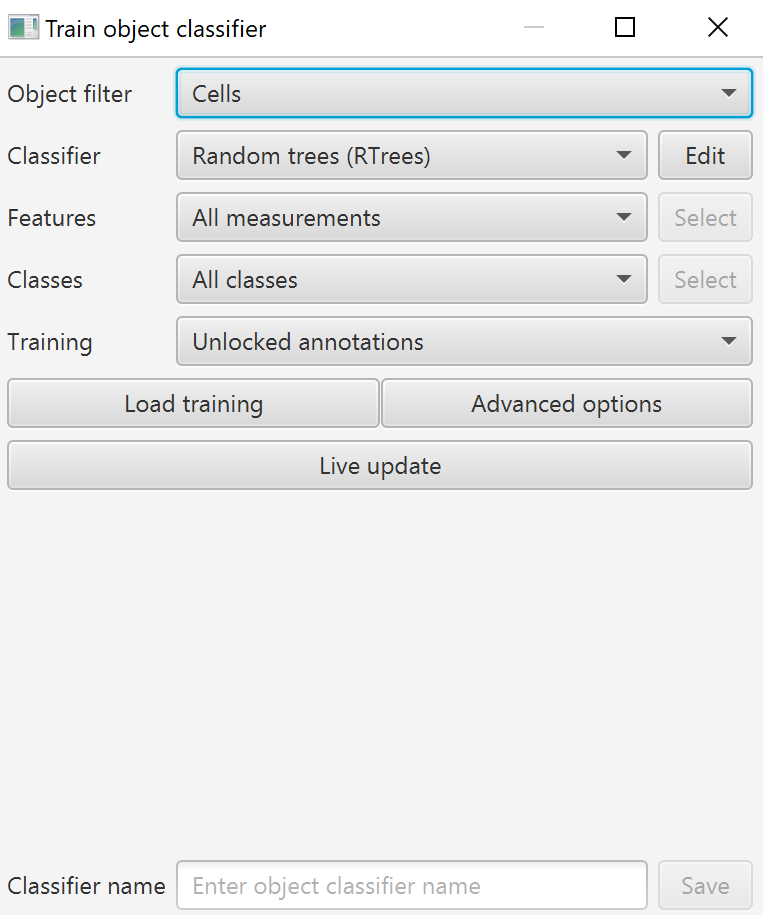
Train object classifier interface
To create a machine learning classifier, we will use Classify->Object classification->Train object classifier. If you run into any issues, Classify->Object classification->Reset detection classifications is a quick way back to blank. However, be aware that it will also reset the class on any subcellular detections you might have, so you may want a script to avoid declassifying your spots..
Object filter: The object filter allows you to choose which kinds of objects you will classify. There is no option to classify annotations. If you want to classify annotations, create them as detections first, add measurements, classify them, then use a script to convert them into annotations of that class. “Cells” are a good option if you want to restrict your classifier to only cell objects, as other types of detections can be created, either by the Pixel classifier, Subcellular detections (think FISH staining, RNAScope), Tiles & superpixels, or other objects manually created by scripts. I would only choose “Detections (no subtypes)” if I specifically want to classify non-cell objects like subcellular detections. Tiles can be useful for specifically selecting SLICS or other superpixels. Probably 95% or more of the time, most users will really only want to choose “Cells” here.
Classifier: There are similar classifier options to the Pixel classifier, and as then I recommend starting with the Random trees option, then if you are not getting sufficiently accurate results, switching to ANN once you have narrowed down the list of inputs (measurements in this case). As before, checking the Edit->Calculate variable importance is the most valuable feature that Random trees provides.
Features: Features are everything that can be found in the measurement list shown when an object is selected in either the Annotation or Hierarchy tabs. The bottom left corner of the screen will show a scrolling list of all measurements “owned” by the selected object. Many of these measurements will be generated by default, but many other measurements can be added through the Analyze->Calculate features menu, including:
Further color/channel based measurements, including texture based measurements (Haralick features). Can be useful if you need to change the color vectors and add additional measurements in cases where you have more than 3 stains.
Smoothed features - collect weighted averages of each measurement from objects within a given radius. This can provide additional context to cell classifiers - “what are the cells nearby like?”
Shape features - mostly included for cells, these can be very useful for detections created by the pixel classifier, annotations, or even subcellular detections if they are being used to detect objects other than ISH spots.
Analyze->Spatial analysis has additional options that can calculate distances between different classifications of cells, distances of cells to some nearby annotation border, or create clusters of similarly classified cells. These do tend to be more useful after the cells are classified, but can be used for “sub-classifications” or “derived classifications.” For example “CD4 positive: Clustered” vs “CD4 positive: Isolated”
Which features are used in the object classifier can be chosen through the “Select” button on the right, which becomes available after changing “All classes” in the dropdown to “Selected measurements”.
 Amount of training data
Amount of training data
Classes: In case you have more classes than you want to use (for example you have multiple sets of detections that are not cells, and only want to classify the vasculature but not the subcellular detections), you can have the classifier only accept training data from certain classes of training objects. Essentially, prevent cross-contamination with other objects in complex projects. Again, the “Select” button on the right becomes available after changing “All classes” in the dropdown to “Selected classes”.
Training: The training data that the classifier uses can be further limited to only pay attention to points objects if there are annotations lying around your image as well. This option could allow you to have classified area/line annotations for a pixel classifier and Points annotations for cells, all in the same training image. Options include All annotations, Unlocked annotations, Points, and Areas (lines do not count as Areas!).
Load training and advanced options: Load training functions in the same way as the pixel classifier - you can use this option to load training data from multiple images. I strongly encourage its use in all cases. Advanced options allows for Feature normalization in case you have extreme differences between your images.
Live update: Once you hit Live update, the classes of your cells or other objects are changed. This is not a preview like many of the other options, here you are actually changing the class of your objects. The intention is that you will continue to add new training data or otherwise adjust the classifier in order to get the results you want, then either save the classifier or finish with the dialog box.
Checking and refining the results
After selecting Live update, I follow a similar procedure to the pixel classifier. I open up View->Show log and try to determine what unnecessary measurements I can eliminate. From the look of the log, it is using pretty much the measurements I would expect to detect elongated cells; all of the top measurements are shape related. As such, I will use the Selected features option to remove all staining information. I do this by clicking the Select button, then clicking Select all in the dialog box that shows up. Next I type “OD” into the filter, and click the Select none button to unselect all Optical Density based measurements. If I delete the text in the Filter, I can see that only the shape measurements remain. I will repeat this process with “Cell”, as the only real shape information I have is the nuclear shape, and I want the classifier to focus on that.
If necessary, hunt around for mis-classified cells and correct them by placing a Point object of the correct class somewhere within the cell. As long as Live update is selected, the classifier will updated automatically.
 An amazing script
An amazing script
Train object classifier interface and log file from View->Show log
Select features dialog box after unselecting OD and Cell features
After clicking Apply, I can see that very few cells changed class, but I can be more confident that a weirdly extreme measurement in other tissue slices will not throw off the classifier. Finally, I need to name (in this case, Elongated cells) and Save the classifier so that I can use that in my script via the Workflow.
runObjectClassifier("Elongated cells");
Finishing the script!
Ok, let’s see now - our script previously ended with the creation of a bunch of sub-Tissue annotations, so we need to include the selection of the correct annotations, the cell detection, and finally the classification of the cells.
//This script is currently designed to only detect tissue over 1 million square microns. //Look at the first number in the createAnnotationsFromPixelClassifier to adjust this behavior //Remove this line if you need to keep objects that already exist in the image clearAllObjects() setImageType('BRIGHTFIELD_H_E'); setColorDeconvolutionStains('{"Name" : "H&E Tile 4", "Stain 1" : "Hematoxylin", "Values 1" : "0.51027 0.76651 0.38998 ", "Stain 2" : "Eosin", "Values 2" : "0.17258 0.79162 0.58613 ", "Background" : " 243 243 243 "}'); //Create the whole tissue annotation createAnnotationsFromPixelClassifier("Tissue", 6000000.0, 50000.0, "SPLIT") selectAnnotations(); //Create sub-tissue regions for the hematoxylin dense, eosin dense, normal, and excluded areas createAnnotationsFromPixelClassifier("Tissue regions", 500.0, 500.0, "SELECT_NEW") //Add cells to the desired regions and classify them selectObjectsByClassification("EosinDense", "NormalTissue"); runPlugin('qupath.imagej.detect.cells.WatershedCellDetection', '{"detectionImageBrightfield": "Hematoxylin OD", "requestedPixelSizeMicrons": 0.5, "backgroundRadiusMicrons": 8.0, "medianRadiusMicrons": 0.0, "sigmaMicrons": 1.5, "minAreaMicrons": 10.0, "maxAreaMicrons": 400.0, "threshold": 0.25, "maxBackground": 2.0, "watershedPostProcess": true, "cellExpansionMicrons": 5.0, "includeNuclei": true, "smoothBoundaries": true, "makeMeasurements": true}'); runObjectClassifier("Elongated cells");
Results from one image.
Due to QuPath using the cell centroid to place a cell “in” a given annotation, some cells at weird edges of annotations will show up as being in the HematoxylinDense areas, despite no cell detection being run there.
RGB version of a section of Tile 4
Optical density view of the same section of Tile 4
Testing out the full script on Tile 4 seems to work just fine! I did adjust the color of “Positive” to Cyan so that it would show up on the Eosin background. Looking at the Show annotation measurements (Grid/Spreadsheet button marked in green above) for the one tile indicates that there may be a higher percentage of elongated cells in the Eosin area than in the normal tissue. But will it hold up? Next let us work on exporting the results in a couple of different ways.