Pixel classifier
Brightfield analyses in QuPath
0.3.0RC2 and beyond will have major improvements to the behavior (speed, memory usage) of the pixel classifier, which should help out significantly on most computers.
Tissue detection (simple thresholder)
Pixel classification (tissue regions)
Overview of the pixel classifier
The pixel classifier is comparatively more powerful than the thresholder, but at the same time more fragile and more likely to exhibit unexpected behavior. The best ways to try to keep the pixel classifier doing what you want:
Balance your classes - class balancing can be enabled in the Advanced options, but that doesn’t make up for actually having enough training examples for all classes.
Balance the examples WITHIN your classes. That means not selecting large swaths of “middle of the tissue” for your Stroma or Tumor pixel classes. If you do that, your classes will be heavily weighted towards what the middle of the tissue looks like, and perform relatively poorly at the edges or in outlier cases.
Try to only include meaningful measurements. The current QuPath interface does not quite make this as easy as a program like ilastik, but do your best to not simply throw all of the measurements at the classifier and hope for the best. Calculating the variable importance using Random Trees can be very helpful here, even if you end up using a different kind of classifier.
Back up your training data
With these steps laid out, we should select a test set of images to perform steps like determining thresholds for positivity. I will use Tile 4.ome.tif and Tile 1.ome.tif as I already noticed that “Tile 4” has a tissue fold on it, which will be useful for the pixel classifier training. “Tile 1” will help ensure a minimum size threshold is set so as not to include extra bits of tissue.
There are two main differences between the pixel classifier and the thresholder (many others as well).
Inputs: There are many, many possible inputs for the pixel classifier, while the thresholder has essentially one.
Training data: Both options let you look at the results in overlays, but only the pixel classifier actually has training data and can pull information from multiple source images.
Training data is important to save, as it is not easy to edit an old pixel classifier. Certain things can be edited in the resulting JSON file, but if you ever want to retrain or “improve” the classifier, you need that original training data. On the other hand, you also want to use your images to run the real analysis on. What to do?
Export the training annotations using a script, or in 0.3.0 as a JSON file.
Duplicate the images within the project. This does NOT take up more hard drive space - you are simply creating a duplicate entry in the project that accesses the same image file.
Duplicate the project. This is by far the safest option. When you get to the stage where you want to create a pixel classifier, make a copy of the project folder, name it after the pixel classifier you intend to use it for, and then create the classifier in that project. Once you are done, transfer the classifier back to the main project (copy and paste the pixel_classifiers folder from the classifiers folder in the training project to the classifiers folder in the real project).
Alternatively, create the pixel classifier in the original project, but then as soon as you are finished creating it, save and close the project, and make a copy of the project folder, renaming it to something about “classifier backup.”
For this guide, we will duplicate the images within the project as that is the easiest to demonstrate, but I encourage you to duplicate the project when handling any important data - the chance of running a script “For Project” without thinking and overwriting all of your training data is too high!
Creating training annotations
To start, create your backup project, duplicate your images, or use whatever method you choose to ensure your training data stays safe. Below the Project tab’s duplicate image feature is used.
Using the Project tab to duplicate an image entry, which does not duplicate the image file itself.
Save your data as you go: The Undo button only works for certain amounts of changing coordinates. Too many big changes and you might find yourself unable to Undo. CTRL+S!
Choose and create classes: Create and recolor your classes before opening the Pixel classifier dialog. I tend to avoid coloring classes white in brightfield images, or black/brown in fluorescent images, but otherwise choose colors and names that make sense to you. Alternatively, choose names that will make sense to whoever is getting your data - ask the biologist or pathologist you are helping. Adding an asterisk after the name will prevent objects from being created from that classification. In this case, I will use Ignore* (default class) for the tissue folds, and WhiteSpace* for empty spaces I would like to exclude from calculations.
Training data needs to be diverse: I am talking about within a single class. One single blob of a particular class might get the general idea, but it will not help the classifier handle edge cases, and borders and edges are frequently what we most want to get right when classifying areas. Taken another way, “purple is one of my classes” is easy for the classifier, so there is no need to bias the training data towards that. A spread of different areas is much better. The amounts of training data in the images below are much higher for the two bad examples than the one good example, even though the good example covers much more tissue area. For those interested, there is an even more detailed description about the use of polylines when training a pixel classifier here.
Bad training data - the majority of it is redundant. Purple with more purple around it.
Mediocre training data. At least there are a variety of areas sampled, but the center of the tissue is greatly over-represented.
A more well rounded training data set. This image is zoomed out and more tissue area is covered, but only polylines are used, so the actual amount of training data is much smaller than the other two examples.
Return to Move tool automatically has been turned off in Edit->Preferences
Generating training data faster: When creating the training data there are a couple of other useful interface tools. The first, is not needing to click the drawing tool you want to keep using every time you want to use it. If you are comfortable drawing and using the keyboard, the shortcut keys like V for the Polyline (the button looks like a V), B for Brush or W for the Wand Tool all help get back in the action quickly. Another nice combination, if you are exclusively using one tool, is to turn off the default-enabled “Return to Move Tool automatically” shown to the right.
Now, if you select the Polyline tool, you will keep drawing polylines every time you left click. But how to move the screen since you will always draw a polyline? Hold down the space bar, and every left click and drag will move the screen regardless of what tool is selected or what is under the cursor. The space bar is a great option when you have large annotations that are not locked, and you want to avoid accidentally moving!
Finally, you can use the Auto set button at the bottom of the Class list in the Annotation tab. If you know you will be creating many annotations of a single class in a row, this will save you having to assign classes to each one. Simply select the class you want to start drawing, select the Auto set button, and start annotating!
Pixel classifier interface
Classifier and Edit: The two main options here are ANN and Random Trees. I prefer Random Trees, and will leave the other two options to others to explain/explore as they have generally been slow and not provided great results. Random Trees is a good way to start due to the Edit menu option, Calculate variable importance. This option, even if you later choose to go with an ANN, gives you insight to the inputs that the classifier prioritizes. This information helps in two ways. First, it allows you to eliminate measurements that are unnecessary, speeding up the performance of the classifier, and possibly improving its generalizability (to new data, by not being overly specific). Second, it provides a sanity check, as you can look at the measurements that are most important and decide whether they make sense.
Resolution: Resolution is fairly straightforward - it is the baseline view at which you are going to analyze the image. Lower resolutions are the equivalent to zooming out, and will be both faster and pick up less detail. If your pixel classifier is intended to measure subcellular objects, it will need to be of the highest resolution. Alternatively, if you are simply detecting tissue, folds, and background, you may want a fairly low resolution. The mid-range resolutions, like what I use for this project, are a middle ground where large scale features are important, but texture measurements are important - measurements that would be unobtainable at low resolution. When defining Features, the filters at 0.5 to 8.0 times the pixel size will be based on the chosen pixel size! In this particular project, if you want to pick up some of the finer collagen fibers, you will need to increase the resolution. When you change the resolution, it is usually a good idea to revisit any Features that were previously excluded, as those features may be more useful at picking up fine details at high resolution.
The Add button allows you to define your own resolution. I have never needed this.
Features: The dropdown here is not terribly important, but the two buttons, Edit and Show are some of the most important in the whole interface.
Show will bring up an ImageJ stack showing every combination of channel, sigma, and feature, so that you can see what inputs are going into your classifier. Aside from using the Random Trees Calculate variable importance, this is the only other way I know of to easily determine the importance of your features. What you are looking for are features that show a strong or consistent difference between the types of regions in your image. The image generated by the Show button will be the region currently in the Viewer that is part of the Pixel classifier window, and the resolution will be determined by the current setting of the… well, Resolution. Finally, Show generates the same sort of output as the Show classification dropdown below the Viewer.
Edit is where you choose all of your inputs to the classifier. I wish this button were highlighted a bit more, but here is where you choose whether to use RGB values, color vectors, low res or high res versions of filters, and a decently large selection of filters.
The features list has a brief explanation in the official documentation, but I strongly recommend investigating for yourself which are important to your project through Random Trees and the Show button
I have not yet tested Local normalization - I prefer to insist on quality input data :) If anyone has feedback on this feature, I would appreciate it, and will give all credit to any contributions either here or on the forum. I will simply quote the official documentation.
“Local normalization: Generally best avoided. This can optionally apply some local background subtraction and normalization in an effort to handle image variations, but in practice it often does more harm than good. May be removed or replaced in a future version.”
Output and Region: Output should mostly be left as Classification, but in some cases you may want to see the Probability to determine where the classifications are “close” rather than “certain.” The colors of the different classes will blur together, so using contrasting colors can be very helpful here. In addition, there will be an additional Prediction line below the pie chart showing class balance showing the relative weights of the predictions of each class in the pixel with the cursor over it. Usually the class names will be shown, but not always. You may have to move the cursor around to get class names to show up.
Load training: As will be mentioned below, the Load training button allows the user to import annotations (and their pixel information) from other images. This option is how you build a diverse and robust pixel classifier.
Advanced options: There is a bit to unpack here, but the one option that I enable about 100% of the time is checking the Reweight samples option.
The Maximum samples has a very direct impact on the speed of training the classifier, though usually less of an impact on generating the actual overlay. Each pixel (at the selected Resolution) under any training annotation is a “sample”. With carefully curated class annotation, this value should rarely be a problem (making the RNG seed unnecessary). However, selecting large swaths of tissue for your training data is where this value comes heavily into play. If you select 500,000 pixels across several images as your training data, by default you will only ever be using a random one fifth of it. That one fifth might not contain any of the edge cases you are concerned about (borders between tumor and stroma, for instance). I am not aware of any straightforward way to calculate the current number of samples in your data set, but it could probably be scripted.
RNG seed: If you have more than the Maximum samples worth of training pixels, adjusting the RNG seed may give you different results for your classifier. The more over the Maximum samples you are, the bigger changes you should expect to see as you adjust the seed.
Reweight samples: The best. Balances your samples for you, to some extent. This option does not replace having a diverse training data set, but for instances where there is a lot of variation within one class, but very little variation in another, it can save you from needing to create many needless annotations just to balance your classes. When exploring the project you might notice that my “Whitespace*” class has far fewer training examples, which I am comfortable with since most of the whitespace is fairly homogenous. As long as I have enough examples of the “edge” of the white space, I should be fine (due to the downsample occurring when picking a Resolution of Moderate, the edge pixels will contain some tissue).
Preprocessing: I have not really used this, sorry, ask on the forum!
Annotation boundaries: Very useful for finding things like nuclei edges if you want to create a pixel classifier to detect nuclei, but not have touching nuclei merge together. By default this is set to Skip and is not used. I would only recommend using this option if you are NOT using linear annotations. One use case might be selecting a “Boundary strategy: Classify as Border” for a new Border class you created, and create outlines of a few nuclei using the Wand tool. With the boundary set to 1 or 2 pixels, you could automatically train the classifier to create an “edge of the nuclei” class, without manually annotating the edges of the cells you are training on.
Live prediction: Live prediction is the button to hit when you want to start inspecting the results of your Pixel classifier options selections. For high resolution classifiers this can be very slow, and in version 0.2.3 and earlier could slow even extremely powerful computers to a crawl. Once you press the button, a pie chart should show up below the button to give you an idea of your current class balance.
Show classification dropdown: The Show classification dropdown menu below the Viewer gives you the ability to look at the entire tissue slice as it would be seen through a given filter. The slider below the dropdown can be used to swap back and forth between the filter view and the original image.
A pair of images showing the Eosin Structural tensor min eigenvalue with a sigma of 4.0, at a Resolution of Moderate.
Inspecting the overlay with the image in the background shows me that this value is picking up regions of fragmented tissue with lots of edges between Eosin and background. It is also picking up regions within the tissue fold.
Checking your results - updating your classifier
Looking at Variable Importance can let you reduce the complexity of your classifier, making it run faster, and potentially be more generalizable to new data (avoid overfitting).
My first step after pressing Live prediction is a visual overview of the results, making use of the Opacity slider or “C” button in the top center of the GUI to look below/through the masks, or toggling the “C” key when the Viewer is in focus (in other words, the “C” key will not do anything if you have the Pixel classifier window selected instead of the image in the Viewer). After looking around the image+mask to make sure nothing is terribly wrong, my next step is to look at View->Show log (shown to the right) in order to determine whether the classifier is selecting good information to make its decisions, and which inputs I might be able to remove to speed up the classifier.
The resulting list shows me that low resolution Hematoxylin and Eosin Gaussian are the most important, which is good! The classifier is picking up that I want broadly Eosin and Hematoxylin stained areas. I also see several min and max Structural Tensor eigenvalues, and the occasional weighted deviation. The sigma values tend towards 4 and 8, but I see enough 0.5, 1 and 2 values to keep them all.
This all means that I can reduce my inputs from 12 features to 4 features, which is a reduction of
8 (features) * 3 (channels) * 5 (sigmas) = 120 inputs.
The final features I keep are Gaussian, Weighted deviation, and Structural Tensor min and max Eigenvalues.
I did notice that there are several areas around the edge of the tissue that are not being picked up. Those look a bit scraggly, and if there were a biologist or pathologist leading the study, I would definitely check with them whether or not to add more training data in order to keep those areas.
Expand the training data set across multiple images
Instead of digging into the biology in this guide, we will move on to Tile 1 (the training image) and see what we get! If you switch images with the Pixel Classifier open and the overlay turned on, the new image will immediately begin to get its own overlay. If you want to quickly move to a new image and begin annotating, I recommend turning either the overlay (“C”) or the Live prediction off.
Browsing around Tile 1 with the Overlay at ~30%, I immediately notice that there is a lot of "blue” where there should not be, indicating that the nuclei dense regions in this image are being treated as HematoxylinDense instead of as NormalTissue. This sample simply has much denser cell populations than my previous sample in Tile 4, so I need more training data. This is a great example of why creating a pixel classifier based on just one image is a bad idea!
Orange arrows indicate some of the problem areas.
Where is my pixel classifier overlay?!
To make QuPath run a little bit more smoothly, I first turn off the Live prediction, and then draw some areas for additional training. The downside is that the overlay disappears when I create a classified annotation, but you can draw all of your new annotations without classifying them (maintaining the overlay) and then assign the classes later. The benefit of turning off the Live prediction is that I do not have to wait for QuPath to recalculate the entire classifier each time I add one annotation.
So I click live prediction after adding a few new line annotations and classifying them as NormalTissue. Nothing happens. What is going on?? Well, the Pixel classifier is only pulling data from the current image, and I only have one class present. I need to load the data from the previous image - which can be confusing since the classifier seemed to work just fine before I created new annotations!
Click the Load training button right above the Live prediction, and include the previous image’s (“Tile 4ome.tif training”) training data. Now, not only does the overlay work, as we now have more than one class of data, the classifier is built off of the training data from the current image and the image you loaded. You may need to reload data each time you move to a new image, if you are classifying across several images.
Back in business, and the training areas seem to have worked to improve the normal tissue classification.
Now that I am happy enough with my “test set” of two images, I save both the training data (my image) and the classifier. With the classifier saved, I can now use the Measure, Create Objects and Classify buttons.
Finishing up and adding to the script
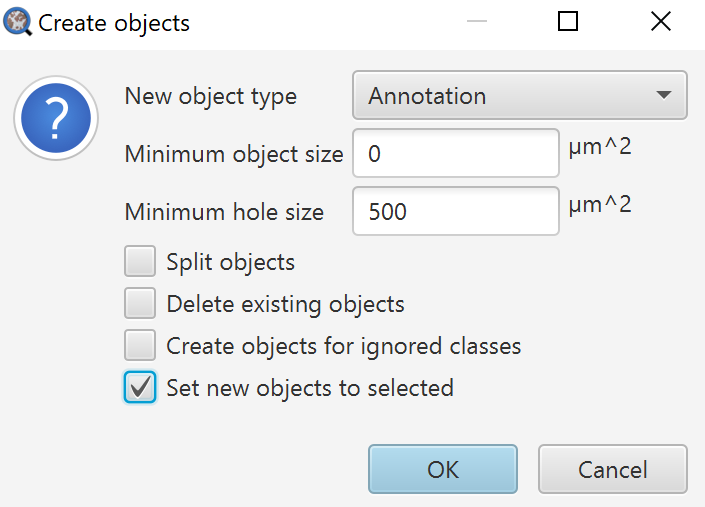
What did we want to do with this again? Right, create cells in certain regions, and compare area measurements. While I could use the Measure button to handle the latter, I already know that each annotation created will automatically include an area measurement, so I can achieve both goals by only using Create objects. The dialog for Create objects is the exact same as in the previous Tissue detection page, so reference that if you need more details on the options. For this use case, I will use:
As I want annotations to create cells within, but I want to avoid really small isolated bits of annotation, so I set a Minimum object size threshold and a Minimum hole size threshold that are the same. While this can result in missing areas along classification borders, in most cases isolated, misclassified pixels will both be removed, and the hole will be filled in, resulting in contiguous and non-overlapping annotations. Try removing the Minimum object size threshold to see what the results are! The resulting code from the workflow is:
createAnnotationsFromPixelClassifier("Tissue regions", 500.0, 500.0, "SELECT_NEW")
I can add that code to my overall workflow, although I do need one additional step, a selectAnnotations(); line to make sure the “Tissue” step annotations are selected - this lets the “Tissue regions” pixel classifier know which areas to work inside of. The current code now looks like the following.
//This script is currently designed to only detect tissue over 1 million square microns. //Look at the first number in the createAnnotationsFromPixelClassifier to adjust this behavior //Remove this line if you need to keep objects that already exist in the image clearAllObjects() setImageType('BRIGHTFIELD_H_E'); setColorDeconvolutionStains('{"Name" : "H&E Tile 4", "Stain 1" : "Hematoxylin", "Values 1" : "0.51027 0.76651 0.38998 ", "Stain 2" : "Eosin", "Values 2" : "0.17258 0.79162 0.58613 ", "Background" : " 243 243 243 "}'); createAnnotationsFromPixelClassifier("Tissue", 6000000.0, 50000.0, "SPLIT") selectAnnotations(); createAnnotationsFromPixelClassifier("Tissue regions", 500.0, 500.0, "SELECT_NEW")
The resulting overlay can be exported manually using File->Export images…->Rendered RGB (with overlays)
The result I have obtained here is certainly not the best possible! More training data, and better balanced training data across more images could certainly help improve the results. I leave plenty of room for improvement to you, the reader!
The above images could also be exported en mass for the project using scripts from the QuPath Readthedocs site as follows:
// Write the full image downsampled by a factor of 10 def requestFull = RegionRequest.createInstance(server, 10) writeImageRegion(server, requestFull, '/path/to/export/full_downsampled.tif')
 Note
Note